Gut Microbial Metabolism of Xenobiotics
Our primary focus is on decoding the complex metabolism of xenobiotics, including drugs and dietary compounds, mediated by the gut microbiome
Selected Publications: Nat. Chem. 2025; Cell 2020

Illustration created using BioRender
Teeming with metabolically active microorganisms, the human gut transforms xenobiotics, including drugs and dietary compounds, ultimately impacting their duration and bioavailability. We develop high-throughput mass spectrometry technologies to comprehensively map these chemical interactions, aiming to uncover their impact on therapeutic outcomes and host physiology.
To achieve this, we employ a multidisciplinary approach that integrates cutting-edge chemistry and biotechnology to explore unconventional chemical interactions between gut microbiota and xenobiotics. Specifically, our goals are to (i) identify key microbial metabolic pathways involved in xenobiotic metabolism, (ii) determine how microbial biotransformations alter therapeutic efficacy and safety, and (iii) investigate the downstream effects of these processes on host physiology using tissue culture and animal models.
Our research focuses on translating discoveries into targeted applications, such as developing molecular probes to monitor gut microbiome-related therapies, and addressing medical challenges tied to microbiome-mediated drug metabolism. By transforming our understanding of microbial xenobiotic metabolism, we aim to identify potential strategies for improving therapeutic efficacy and patient outcomes.
Ultimately, our work will benefit the pharmaceutical industry in drug development and design, optimized clinical efficacy, and personalized medicine initiatives.
Paired Omics-Directed Natural Product Discovery
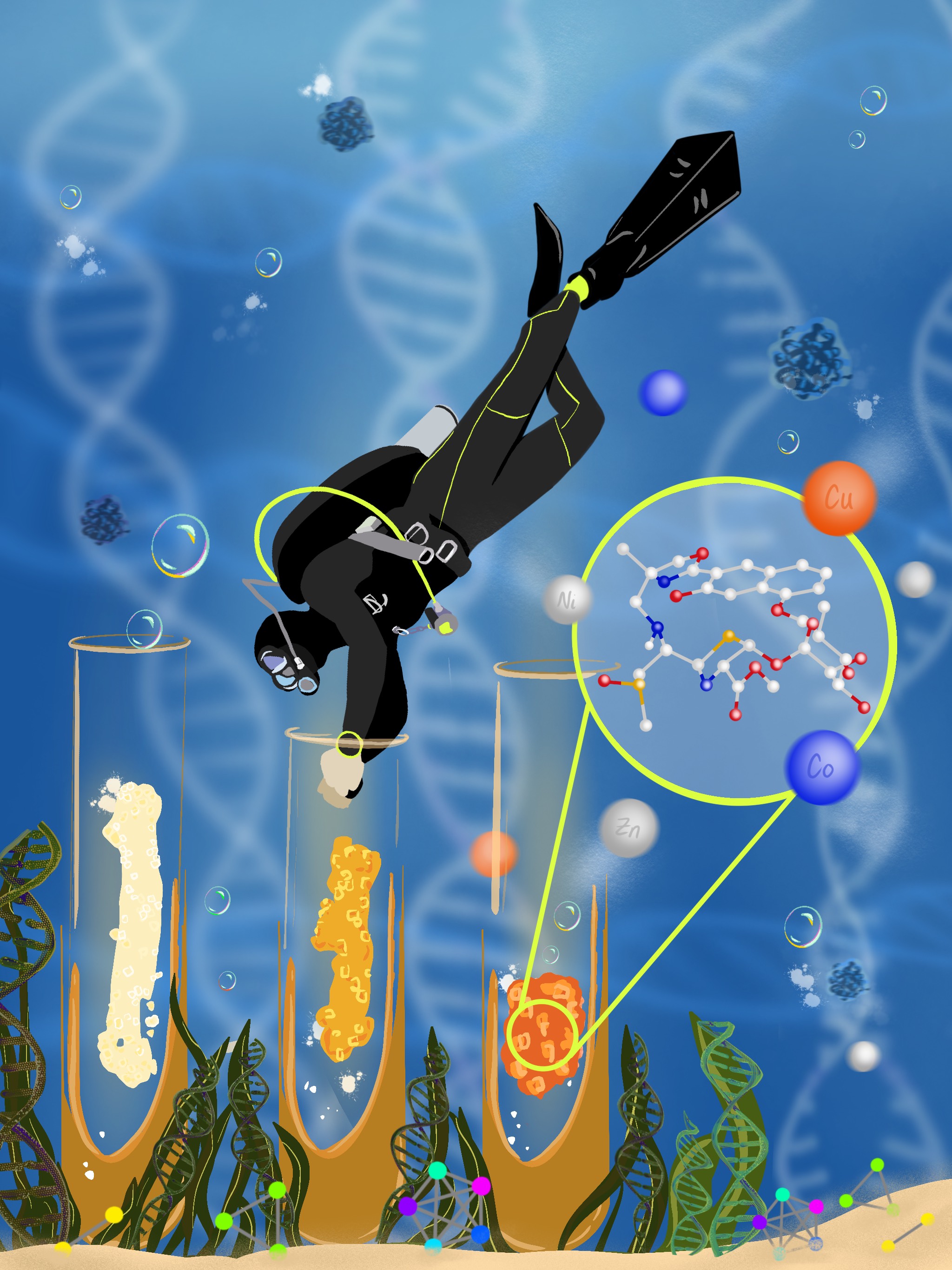
We leverage “omics” technologies to unlock the untapped potential of “orphan” biosynthetic gene clusters from marine and human microbial symbionts for drug discovery and development
Selected Publications: Cell Chem. Biol. 2025; JACS 2025b; JACS 2025a;JACS 2023; ACIE 2020
Although microbial genomes harbor an abundance of biosynthetic gene clusters (BGCs), significant technological gaps remain that hinder the direct correlation of newly discovered gene clusters with their corresponding secondary metabolite products. To address these challenges, we aim to develop a systematic platform integrating advanced omics technologies, enabling the discovery and activation of orphan BGCs, thereby providing access to untapped natural product resources.
By investigating marine and human microbial symbionts we combine paired omics approaches (metabolomics and genomics) to activate orphan BGCs. This platform facilitates the discovery of bioactive secondary metabolites, including potential leads for antibiotic and anti-cancer drugs. As a proof of concept, we discovered a new class of nonribosomal peptidic metallophores from a marine sponge bacterial symbiont, uncovering the novel enzymology underlying biosynthetic assembly and the role of cluster collaborations or ‘duets’ in producing such structurally complex agents. Our integrated approach combines genome mining, metabolomics, and activity profiling to simultaneously target structural novelty and biological activity in natural products.
This strategy enhances the efficiency of small molecule drug discovery, enabling a more targeted exploration of previously uncharted chemical space.